Article highlights
-
Epigenetics refers to the heritable changes in DNA expression without changes in the genetic code.
-
Epigenetic changes are brought about by post-translational modifications of histone proteins, covalent modifications of DNA bases and microRNA.
-
Epigenetics explains how environmental milieu such as diet, physical activity, circadian rhythm, intrauterine malnutrition or maternal obesity interact with the genome of an individual and lead to diseases such as type 2 diabetes mellitus (T2DM).
-
Epigenetics also contributes substantially to the development of micro– and macrovascular complications of T2DM.
-
Current research to develop newer drugs that target the epigenetic dysregulation in T2DM is ongoing.
Diabetes has become a global pandemic, with an estimated 536.6 million people living with diabetes worldwide in 2021, and this is likely to increase to 783.2 million by the year 2045.1 The primary pathophysiology of type 2 diabetes mellitus (T2DM) involves insulin resistance in the liver, adipose tissue and skeletal muscle, followed by defects in insulin secretion later in the course of the disease.2 Notably, T2DM is a polygenic disorder that develops complex interactions between genes and the environment.
Epigenetics refers to heritable changes in DNA expression without alterations in the genetic code.3 In the past few decades, understanding the epigenetics of T2DM has unravelled the missing pathogenic links in the causation of the disease.4 Simultaneously, it has also enabled us to understand the effects of environmental factors, such as diet and physical activity, on the pathogenesis of the disease. Epigenetic changes can be used as potential biomarkers to assess the risk of the onset of T2DM and vascular complications. Epigenetic alterations can also predict the response to therapy and lifestyle interventions, thereby offering a tool for precision medicine.4
With the ever-increasing knowledge of epigenomics,5 it is not possible to review all epigenetic mechanisms of T2DM comprehensively. In this review, we discuss the basic mechanisms of epigenetics and try to summarize the important epigenetic dysregulation implicated in pathogenesis, vascular complications and therapeutics to appreciate its impact on our current understanding of T2DM.
Basics of Epigenetic mechanisms
In the 1940s, the term ‘epigenetics‘ was used to refer to the complex interactions between the genome and the environment.6 This concept has evolved significantly in the past 50 years. Riggs et al. defined epigenetics as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in the DNA sequence”.3 Epigenetics-mediated differential gene expression can explain the heterogeneous functions of different cell types in the body despite them all carrying the same genetic information.6 It is now known that epigenetic changes also control non–coding regions of DNA, which are essential in various physiological and pathophysiological states.7 Epigenetic changes can be classified into two types: direct epigenetics and indirect epigenetic, which is further subdivided into within indirect epigenetics and across indirect epigenetics.8 Direct epigenetics refers to the changes in gene expression occurring in an individual’s lifespan due to interactions with the environment. Within indirect epigenetics refers to the changes in gene expression that occur within the intrauterine environment. Finally, across indirect epigenetics refers to altered gene expression due to epigenetic changes inherited from ancestors.8 Hence, epigenetic changes can influence an individual’s genomic expression from when they are in the zygote state and throughout their whole lifespan, both as a ‘static expression’ (i.e. inherited from their ancestors) and a ‘dynamic expression’ (i.e. due to interactions with the environment).
The process of protein synthesis takes place in two steps: transcription, which is the synthesis of messenger RNA (mRNA) by copying a gene’s DNA sequence, followed by translation, in which the information carried by mRNA is decoded to produce peptides by ribosomal RNA and transfer RNA. Epigenetics-mediated altered DNA expression can occur at the level of both transcription and translation. Covalent modifications of DNA bases (methylation) and modifications of histone proteins alter DNA expression at the level of transcription, whereas non–coding RNAs (specifically microRNAs [miRNAs]) affect the gene expression at the level of translation (Figure 1).8,9
Figure 1: Mechanism of epigenetic changes
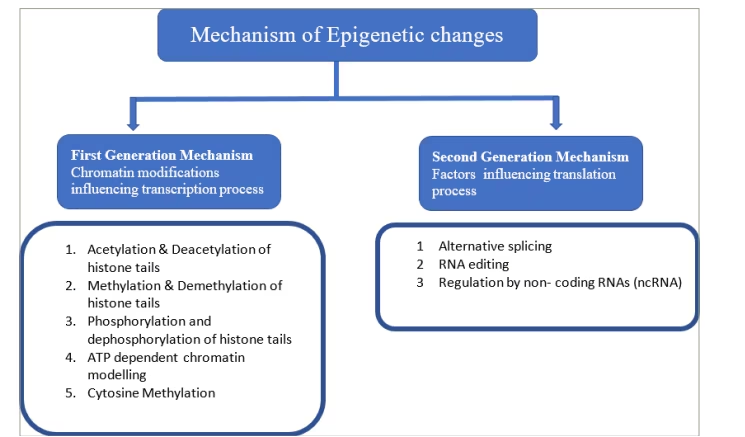
ATP = adenosine triphosphate.
DNA methylation is an enzymatic process by which a methyl group is covalently added to cytosine residues to alter the gene expression. It is carried out by enzymes belonging to the families of DNA methyl transferases (DNMT), namely DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L.8–10 S-adenosyl methionine (a methyl donor) donates methyl groups to cytosine residues in CpG, or cytosine guanine (CG), dinucleotide sites. In humans, most of the promoter sites of DNA have a CpG island that is the target of these enzymes. The hypermethylation of the CpG sites of these target promoters prevents the process of transcription, thus silencing gene expression (Figure 2).11
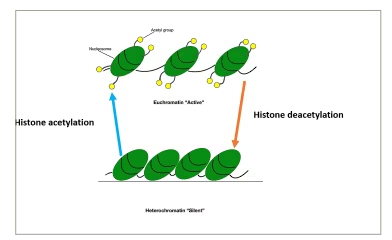
Histone proteins can be modified by methylation, acetylation, ubiquitination or phosphorylation.12 These modifications lead to changes in the structure of chromatin, causing the formation of either euchromatin, which results in gene expression, or heterochromatin, which results in gene silencing.13 Histone methylation can lead to the activation or repression of gene expression. Histone acetylation leads to a decrease in the positive charges of histone proteins, which in turn causes a decrease in its interactions with DNA and increased accessibility of transcription complexes to DNA (euchromatin), leading to increased gene expression. The opposite happens when deacetylation takes place (Figure 3).13
Figure 2: Methylated and unmethylated CpG islands of DNA causing gene repression and expression, respectively
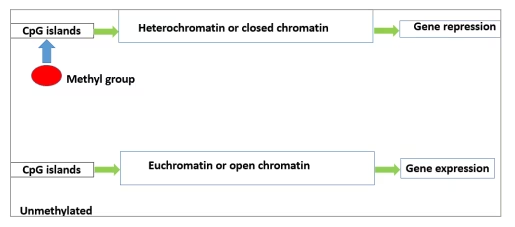
Figure 3: Histone acetylation causing the formation of euchromatin (active chromatin) and deacetylation causing the formation of heterochromatin (silent chromatin)
miRNAs are specialized non–coding RNAs about the size of 22 nucleotides in length.14 They are synthesized by DNA with the help of RNA polymerase II. From the nucleus, they reach the cytoplasm to bind to a specific target mRNA, resulting in the cleavage of bound mRNA causing translational repression (Figure 4).14 MiRNAs can be regulated by processes of DNA methylation, RNA modification and histone modification.
Figure 4: Mechanism of microRNA causing cleavage of target messenger RNA, leading to translational repression
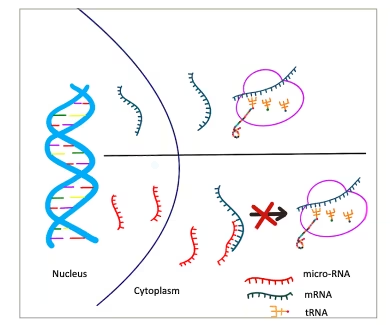
miRNA = microRNA; mRNA = messenger RNA; tRNA = transfer RNA.
Epigenetics and type 2 diabetes mellitus
Epigenetics and risk of type 2 diabetes mellitus
The role of epigenetics in the onset of T2DM can be exemplified by studies showing an increased prevalence of diabetes in adulthood when there is a history of poor maternal nutrition or diabetes in the mother during gestation.15,16 It should be noted that both maternal malnutrition and gestational diabetes put the child at future risk of developing T2DM. Creating an undernutritional environment in utero, especially in early gestation, was hypothesized to lead to the epigenetic programming of various metabolic pathways in the foetus in anticipation of an adverse environment later in life.16 When these children are postnatally exposed to an abundance of nutrition, they are at risk of developing T2DM, obesity and metabolic syndrome. This phenomenon is referred to as the ‘thrifty phenotype’ hypothesis. It has been proposed that maternal malnutrition leads to decreased levels of leptin in the blood, which may later lead to obesity.16 Studies of the Dutch Hunger Winter famine cohort found that children exposed to famine in early gestation showed a 5.2% decrease in DNA methylation of the insulin-like growth factor 2 (IGF2) gene differentially methylated region compared with children who had normal maternal nutrition in pregnancy or were exposed to famine during the later part of pregnancy.17 Apart from these methylation changes, increased expression of miR-576–5p seems to play an important role in the pathogenesis of increased cardiometabolic risk.17 Furthermore, lower circulating levels of miR-15a, miR-29b, miR-126 and miR-223 and higher levels of miR-28–3p can predict the risk of developing T2DM.18
During postnatal life, factors such as diet, physical activity, sleep–wake cycle and various environmental factors lead to changes in the epigenome and contribute to the risk of developing T2DM in the future.15 An important RNA-binding protein, NONO, is an epigenetic regulator of genes, controlling various pathways of carbohydrate and fat metabolism in the liver in accordance with the availability of nutrition.19 NONO is pivotal in predicting an individual’s risk of developing T2DM.15,19
Yajnik depicted the thin-fat Indian phenotype in an article where he describes that Indian babies are born with low birth weight but are found to have higher visceral fat than their English counterparts and are predisposed to insulin resistance and T2DM in later life.20 The Developmental Origin of Health and Disease theory traces the origin of T2DM to intrauterine life followed by rapid childhood growth leading to a biphasic nutritional insult.21 Yajnik et al. have also reported that nutritional factors such as 1–C (methyl) metabolism with normal to high maternal folate levels and vitamin B12 deficiency in mothers predisposed Indian babies to higher adiposity and insulin resistance by foetal epigenetic changes.22
The complex interactions of epigenetics in the evolution and progression of T2DM are summarized in Figure 5.15
Figure 5: Epigenetics and type 2 diabetes – complex interactions at various levels
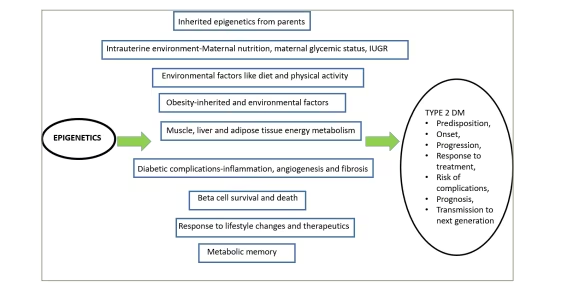
IUGR = intrauterine growth restriction; T2DM = type 2 diabetes mellitus.
The epigenetics and pathogenesis of type 2 diabetes mellitus
Two primary components in the pathogenesis of T2DM are insulin resistance and impaired insulin secretion. Various studies have demonstrated altered DNA methylation of genes, namely PPARG, KCNQ1, TCF7L2 and insulin receptor substrate 1 (IRS1), that are involved in the actions of insulin at sites such as the liver, skeletal muscle and adipose tissue.23–31 Pancreatic islets of patients with T2DM have shown decreased expression of genes involved in insulin secretion, such as PPARGC1A, INS and PDX1, due to increased methylation.32–34 Decreased expression of PPARGC1A is also noted in the skeletal muscles of subjects with T2DM.35 Further human and animal studies of skeletal muscle suggested methylation defects in genes that regulate insulin sensitivity, such as NDUFB6,36 COX5a,37 OXPHOS,38 PGC-1α,39 PDK4 and PPAR-δ.40 Studies in rats with obesity induced by a high-fat diet suggested increased methylation and, subsequently, decreased expression of glucokinase and L-type pyruvate kinase promoter regions, which are involved in the pathogenesis of insulin resistance in the liver.41 Upregulation of histone deacetylase 7 (HDAC7) in human pancreatic cells of individuals with T2DM was found to be associated with a decrease in glucose-mediated insulin secretion.42 Acetylation of the FOXO1 gene, which controls PDX1, leads to an impact on beta-cell development and glucose homeostasis.43 HDAC6-mediated histone3 lysine 9 (H3K9) deacetylation leads to the downregulation of the IRS2 protein, which in turn leads to the development of insulin resistance.44 MiRNAs were found to be important in the processes of beta–cell dysfunction and beta–cell survival, both of which are crucial events in the pathogenesis of T2DM.45 The miRNA MiR–375 participates in the development of the pancreas and decreases insulin secretion by inhibiting myotrophin.46 Another miRNA, miR–124a, impacts insulin secretion with a mechanism similar to that of miR-375.47 MiR-29a and MiR-29b inhibit the secretion of insulin by their inhibitory actions on monocarboxylate transporter 1 (MCT1).48 MiR-184 can induce beta–cell replication, thus causing an increase in the beta–cell population.49 MiRNAs also contribute to the pathogenesis of insulin resistance by their action on the phosphoinositol-3 kinase (PI3K)/AKT and other insulin signalling pathways.50 The following MiRNAs regulate these proteins: miR-128a, miR-96 and miR-126, which control the expression of the IRS-1; miR-29, miR-384–5p and miR-1, which regulate PI3K expression; miR-143, miR-145, miR-29, miR-383, miR-33a, miR-33b and miR-21, which modulate AKT expression; and miR-133a, miR-133b, miR-223 and miR-143, which control the expression of the glucose transporter GLUT4.51
Epigenetics and complications of type 2 diabetes mellitus
Metabolic memory
Metabolic memory, or legacy effect, refers to the benefits of early good glycaemic control to the overall positive effects in the course of T2DM.52,53 Good glycaemic control early in the natural history of T2DM leads to long-term protection from micro– and macrovascular complications irrespective of the glycaemic status in the later part of the disease.52,53 On the other hand, due to metabolic memory, initial poor glycaemic control may lead to a higher risk for vascular complications in the later course of the disease.52 The evidence for this effect comes from three large, randomized controlled trials with long-term follow-up data, namely the Diabetes control and complications trial in type 1 diabetes,54 the United Kingdom prospective diabetes study55 and Steno-2 in T2DM.56 In these studies, patients who were in the initial intensive arm continued to show decreased incidence of vascular (both micro and macro) complications on long-term follow-up compared with patients who were initially in the conventional arm and later switched to intensive therapy despite presenting similar glycaemic status in the later phases.
The basic pathophysiological mechanisms of metabolic memory are hyperglycaemia-induced damage to mitochondrial DNA and its proteins, stimulation of protein kinase C, activation of sorbitol pathway and formation of advanced glycation end products.57 It has been proposed that hyperglycaemia leads to permanent epigenetic modifications of DNA expression of the abovementioned pathways that are consistent with poor glycaemic states, and they continue to persist even after good control is achieved at a later stage (Figure 6).58 Epigenetic mechanisms include changes in post-translational histone modifications, DNA methylation and miRNAs leading to irreversible changes, referred to as metabolic memory.59
Figure 6: Mechanism of ‘legacy effect’
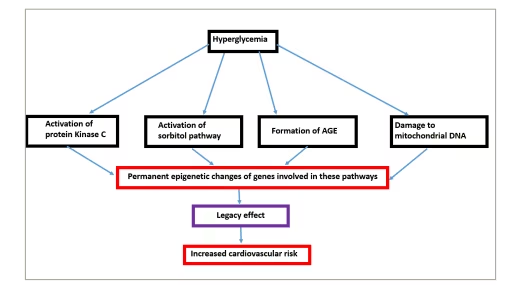
AGE = advanced gycation end product.
Endothelial dysfunction
Endothelial dysfunction seems to be a crucial component in the development of all vascular complications of diabetes. Chronic, uncontrolled hyperglycaemia leads to vascular damage by multiple pathways, namely oxidative stress, increased production of advanced glycation end products, activation of inflammatory and fibrotic pathways by transforming growth factor-beta (TGF-β), nuclear factor κβ (NF-κβ) and angiotensin II (AngII).15 Endothelin 1 (ET-1), a peptide produced by the vascular endothelium, causes vasoconstriction and increased fibrosis, and is known to be abundant in patients with vascular complications in T2DM.60 Methylation defects are noted in the CpG regions of the promoter of the EDN1 gene, which leads to its overexpression.59 Decreased methylation and increased acetylation of histone proteins in the NF-κB gene promoter region leads to overexpression of this proinflammatory marker in the endothelial cells.61 Increased acetylation of histone H3K9/K14 leads to the overexpression of other inflammatory markers, such as interlukin-8 (IL–8) and heme oxygenase gene 1 (HMOX1), in the endothelial cells of the aorta.61 Hyperglycaemia affects H3K9 demethylation, leading to increased expression of matrix metallopeptidase-9 (MMP-9), which in turn causes damage to the mitochondria and endothelial cell death.62 MiR-140–5p, miR-221–3p, miR-200b and miR-130b-3p participate in the pathogenesis of endothelial dysfunction by targeting several genes related to apoptosis, inflammation, hyperpermeability, senescence and pathological angiogenesis.63 Apart from the these microRNAs, miR-126 overexpression is associated with endothelial dysfunction of peripheral arterial disease in T2DM.64
Macrovascular complications
DNA methylation defects are noted in genes involved in the formation of atherosclerotic plaques, such as SOD2, FGF2, ABCA1, COX2 and SMAD 7.65 DNA demethylation causing overexpression of the KLF4, KLF5 and OPN genes leads to the increased mitotic activity of smooth muscle cells in coronary vasculature.66 Hyperacetylation of histone proteins H3K9 and H3K27 has been implicated in stabilizing atherosclerotic plaques.67 Moreover, alteration in histone proteins associated with oxidized low-density lipoprotein–mediated inflammatory response is crucial in the pathogenesis of coronary artery disease (CAD) in T2DM. In one study, miR-126 levels were found to be inversely correlated with serum low-density lipoprotein in T2DM patients with underlying CAD compared with T2DM without CAD.68 Two important miRNAs, namely miR-1 and miR-133, have been shown to be significantly correlated to the risk for CAD in T2DM, such that they can be considered biomarkers for macrovascular complications.69,70 Other miRNAs, such as miR-210,71 miR-2172 and miR-370,73 are also shown to be associated with T2DM-related CAD. Altered levels of miR-451a,74 miR-195–5p75 and miR-146a76 are associated with cerebrovascular events in T2DM.
Microvascular complications
Diabetic retinopathy
Diabetic retinopathy (DR) is an important cause of blindness in adults worldwide and its presentation spectrum can range from non–proliferative retinoapthy (mild, moderate and severe) to proliferative retinopathy, which is associated with new vessel formation and haemorrhages. Both proliferative and non–proliferative DR can be associated with macular oedema, which can further cause severe morbidity. Global DNA methylation seems to be increased early in the course of the development of DR but does not increase further with disease progression.77,78 Animal studies have demonstrated the contribution of hypermethylation of mitochondrial DNA and DNA polymerase gamma to the pathogenesis of DR.79–81 The modification of histone proteins seems to be associated with neuronal cell death and increased vascular permeability of retinal vessels.82,83 Methylation of the histone protein H3K9 by the histone methyl transferase encoded by SUV39H2 is associated with DR onset.84 Increased H3 histone acetylation is noted in animal models of DR linked to activation of the NF-kβ inflammatory pathway.85 In uncontrolled T2DM, alterations in histone proteins lead to MMP-9 overexpression in retinal capillaries, leading to mitochondrial dysfunction and cell death.86 Wide arrays of miRNAs take part in the initiation and progression of DR. MiR-126 regulates the expression of vascular endothelial growth factor 1 (VEGF1) and other vascular adhesion molecules, which are of considerable importance to the pathogenesis of proliferative DR.87 There is decreased expression of miR-31 and miR-184, which in physiological states inhibit new vessel formation.88 Increased expression of miR-21 is of paramount importance in the pathogenesis of DR by contributing to endothelial dysfunction.89 Circulating miRNAs can be used as biomarkers for the early and late complications of DR. One example of such miRNA is miR-210, the levels of which are higher in individuals with DR than in those without.90 Furthermore, the levels of miR-210 are much higher in proliferative DR than in non-proliferative DR.
Diabetic nephropathy
Diabetes–related kidney disease is the leading cause of chronic kidney disease worldwide and can present both with or without albuminuria. Genome-wide studies have shown that increased DNA methylation is correlated with inflammation in diabetic nephropathy.91,92 In animal models of T2DM nephropathy, hypermethylation of the promoter region of Ras protein activator like 1 (RASAL1) has been noted.93,94 Increased expression of transforming growth factor-beta 1 (TGF–β1) in diabetic kidney disease leads to hypermethylation of RASAL-1 causing activation of Ras-GTP signalling.95 This mechanism results in collagen deposition and fibrosis, which is an important step in the pathogenesis of diabetic nephropathy96,97 Altered cytosine methylation in the promotor regions of the mammalian target of rapamycin (mTOR) is conducive to the inflammation of nephropathy.95 Another study showed decreased expression of the transcription factor Krüppel-like factor 4 (KLF4), leading to the hypermethylation of the nephrotic syndrome type 1 (NPHS1) gene, which encodes nephrin, podocyte cell death and albuminuria.98,99 Decreased methylation of the myoinositol oxygenase (MIOX) gene is associated with diabetic nephropathy progression by increasing oxidative stress and fibrosis.100 Alteration of histone proteins leads to increased TXNIP gene expression, causing increased inflammation of mesangial cells.101 High levels of expression of HDAC4 in diabetic nephropathy promote inflammatory changes by inhibiting the STAT1 pathway.102 Mouse models of streptozotocin-induced albuminuria have shown reduced expression of Sirtuin 1/silent information regulator 1 (SIRT1), a histone deacetylase, which leads to decreased expression of claudin–1 in podocytes.103,104 MiR-133b, miR-199b,105 miR-23a106 and miR-30e107 contribute to renal fibrosis, whereas miR-146a is associated with the activation of inflammatory pathways.108 Furthermore, the urinary exosomal miRNA can potentially be used as a biomarker for the onset of diabetic nephropathy.109 For example, increased urinary levels of miR-1915–5p,110 miR-877–3p,111 miR-192 and miR-215 are found in patients with T2DM with diabetic nephropathy.112
Diabetic neuropathy
Diabetic neuropathy is the most common microvascular complication of diabetes, is due to involvement of long nerve fibres and usually presents as distal symmetric polyneuropathy. Decreased DNA methylation of the whole genomic DNA in white blood cells is a potential biomarker for diabetic neuropathy.113 DNA methylation defects have been seen across many genes coding for proteins affecting multiple steps in the pathogenesis of diabetic neuropathy, such as axon guidance, glycerophospholipid metabolism and mitogen-activated protein kinase (MAPK) signalling pathways.114 Hypermethylation of the NINJ2 gene and its subsequent decreased expression have been observed in diabetic neuropathy.115 This protein, expressed in Schwann cells, is essential for the regeneration of peripheral nerves after injury.115 Increased expression of miR-199a3p, which causes the downregulation of the extracellular serine protease inhibitor E2 (Serpin E2), has been attested in the peripheral blood of patients with diabetic neuropathy.116 Decreased expression of miR-25,117 miR-146118,119 and miR-190a5p120 has been noted in animal models of diabetic neuropathy. This results in the modulation of oxidative stress, interleukins and various other inflammatory processes resulting in neuronal injury.121 Other miRNAs significant to the pathogenesis of diabetic neuropathy are miR-128a, miR-155a and miR-499a, which can potentially be used as biomarkers for the screening of diabetic neuropathy.122
Diabetic cardiomyopathy
Diabetic cardiomyopathy is defined as presence of cardiac dysfunction in patients with diabetes in the absence of any other explainable causes such as CAD, valvular heart disease or hypertenion. Epigenetic changes lead to the overexpression of genes of the renin–angiotensin–aldosterone system (RAAS) axis, which is crucial in the pathogenesis of diabetic cardiomyopathy.123–125 Diabetic cardiomyopathy is associated with hypermethylation of the protein sarcoplasmic/endoplasmic reticulum calcium-ATPase 2a (SERCA2a), which is physiologically important for cardiac muscle relaxation.126 This decreased expression of SERCA2a can explain the diastolic dysfunction seen in diabetic cardiomyopathy.123 MiRNAs are pivotal in various steps of the pathogenesis of cardiomyopathy, such as muscle hypertrophy,127 fibrosis,128 mitochondrial dysfunction,129 cell death130 and foetal genetic programming.131 Foetal genetic reprogramming involves decreased expression of the alpha myosin heavy chain (α-MHC) gene and increased expression of the beta myosin heavy chain (β-MHC) gene, which contribute to the development of diabetic cardiomyopathy.131 Altered expression of some miRNAs – namely miR-1,132 miR-146a,133 miR-133a,134 miR-150,135 miR-200c,136 miR-152–3p,137 miR-26a/b-5p,138 miR-29b-3p139 and miR-223140 – is crucial in the hypertrophy and fibrosis of cardiac muscles.
Epigenetics and therapeutics of type 2 diabetes mellitus
Understanding epigenetics has led to the possibility of using epigenetic changes as biomarkers for predictjng the risk of developing T2DM, its complications and for the development of therapeutic targets. As discussed above, many miRNA levels can be used as biomarkers of microvascular and macrovascular complications. There is evidence to suggest that inhibition of specific miRNAs that are involved in the loss of beta–cell function or beta–cell death can result in the improvement of beta–cell functions.45 Histone deacetylase inhibitors can improve insulin sensitivity by increasing the acetylation of lysine amino acids in the insulin receptor substrate 2 (IRS2) protein.141 Apabetalone, a new drug, acts by blocking histone interactions with DNA; this action has been shown to prevent the rise in inflammatory proteins and the development of atherosclerotic plaques.142 Supplementation with lactobacillus can cause changes in the histone methylation profile and improve insulin resistance.143 Treatment of T2DM with metformin causes reduced DNA methylation of genes coding for metfomin transporters, leading to their increased expression and, thus upregulating the beneficial effects on glycaemic control and insulin resistance.144,145 Metformin also causes alteration in the expression of various histone methyl transferases and SIRT1 (deacetylase).144 Glucagon-like peptide 1 (GLP1) receptor analogues, which are currently used for managing T2DM, help preserve beta–cell function by histone modifications and reactivation of pdx-1 transcription.146,147 GLP1 receptor analogues also lead to improvements in the fatty liver, which is mediated by modulation of SIRT1 (deacetylase) and decrease in the expression of NF‐κB.148
New–onset diabetes that occurs during statin therapy is postulated to be due to DNA methylation defects leading to the dysregulation of ATP-binding cassette subfamily G member 1 (ABCG1).149
MiRNA-based therapeutics aimed at targeting various levels of the pathogenesis of T2DM are being tested in animal studies.150–154 MiRNA inhibitors can be used to suppress the overexpression of pathogenic miRNA, and miRNA mimics can be used for overcoming the underexpression pathology.155,156 Antisense oligonucleotides can also be used to modulate miRNA expression.
Conclusions
This review summarizes the evidence supporting the substantial contribution of epigenetics to the pathogenesis and complications of T2DM. Epigenetic modifications start in the intrauterine environment and continue throughout an individual’s life. Epigenetics influences the transmission of T2DM across generations. It also explains how adverse environmental milieus such as food habits, sedentary lifestyle, circadian rhythm, maternal malnutrition or maternal obesity interact with an individual’s genome, leading to various disease states such as T2DM. In addition, epigenetics is instrumental in developing various micro– and macrovascular complications of T2DM. Research is being conducted to develop epigenetic biomarkers that predict the risk of T2DM and its vascular complications. Newer drugs under development aim to correct the epigenetic dysregulation in T2DM. However, further research is required to identify the epigenetic regulators specific to T2DM before novel therapies addressing the pathogenesis and complications of T2DM can be developed.