Wounds on the feet, known as diabetic foot ulcers (DFUs), are a major complication of diabetes. DFUs can become infected, leading to amputation of the foot or lower limb. Patients who undergo amputation experience significantly reduced survival rates.1 In previous studies, researchers have achieved high accuracy in the recognition of DFUs using machine learning algorithms.2–5 Additionally, researchers have demonstrated proof-of-concept in studies using mobile devices for foot image capture and DFU detection.6,7 However, there are still gaps in implementing these technologies across multiple devices in real-world settings.
The Diabetic Foot Ulcers Grand Challenge 2020 (DFUC 2020) is a medical imaging classification competition hosted by the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2020.8 The goal of DFUC 2020 is to improve the accuracy of DFU detection in real-world settings, and to motivate the use of more advanced machine learning techniques that are data-driven in nature. In turn, this will aid the development of a mobile app that can be used by patients, their carers, or their family members, to help with remote detection and monitoring of
DFU in a home setting. Enabling patients to engage in active surveillance outside of the hospital will reduce risk for the patient and commensurately reduce resource utilization by healthcare systems.9,10 This is particularly pertinent in the current post-COVID-19 (coronavirus disease 2019) climate. People with diabetes have been shown to be at higher risk of serious complications from COVID-19 infection;11 therefore, limiting exposure to clinical settings is a priority. The aim of this work is to provide the research community with the first substantial publicly available DFU dataset with ground truth labelling. This will promote advancement in the field, and will lead to the development of technologies that will help to address the growing burden of DFUs.
Related work
Recent years have attracted a growth in research interest in DFU due to the increase of reported cases of diabetes and the growing burden this represents on healthcare systems. Goyal et al. trained and validated a supervised deep learning model capable of DFU localization using faster region-based convolutional neural network (R-CNN) with Inception v2.7 Their method demonstrated high mean average precision (mAP) in experimental settings. However, this experiment used a relatively small dataset of 1,775 DFU images, with a post-processing stage required to remove false positives. Hence, the study is inconclusive for practical use of the proposed method in real-world settings. Improved object detection methods have emerged since this work, such as the very recently proposed EfficientDet, which may provide superior accuracy.12
Wang et al. created a mirror-image capture box to obtain DFU photographs for serial analysis.13 This study implemented a cascaded two-stage support vector machine classification to determine DFU area. Segmentation and feature extraction was achieved using a super-pixel technique to perform two-stage classification. One of these experiments included the use of a mobile app with the capture box.14 Although the solution is highly novel, the system exhibited a number of limitations. A mobile app solution is constrained by the processing power available on the mobile device. The analysis requires physical contact between the capture box and the patient’s foot, presenting an unacceptable infection risk. Additionally, the sample size of the experiment was small, with only 65 images from real patients and hand-moulded wound models.
Brown et al. created the MyFootCare mobile app, used to promote patient self-care via personal goals, diaries and notifications.15 The app maintains a serial photographic record of the patient’s feet. DFU segmentation is completed using a semi-automated process, where the user manually delineates the DFU location and surrounding skin tissue. MyFootCare has the ability to automatically take photographs of feet by placing the phone on the floor. However, this feature was not used during Brown et al’s experiment,15 so its efficacy is unknown at this stage.
Current research in automated DFU detection using machine learning techniques suggests that the development of remote monitoring solutions may be possible using mobile and cloud technologies. Such an approach would help to address the current unmet medical need for automated, non-contact detection solutions.
Methods
This section discusses the DFU dataset, its expert labelling (ground truth), baseline approaches to benchmark the performance of detections and submission rules used in the DFUC 2020 challenge assessment methods.
The DFUC 2020 dataset is publicly available for non-commercial research purposes only, and can be obtained by emailing a formal request to Moi Hoon Yap: m.yap@mmu.ac.uk All code used for the research in this paper can be obtained from the following repositories:
- Faster R-CNN: https://github.com/tensorflow/models/tree/master/research/object_detection
- YOLOv5: https://github.com/mihir135/yolov5
- EfficientDet: https://github.com/xuannianz/EfficientDet
Dataset and ground truth
Foot images displaying DFU were collected from Lancashire Teaching Hospitals over the past few years. Three digital cameras were used for capturing the foot images: Kodak DX4530 (5 megapixel), Nikon D3300 (24.2 megapixel) and Nikon COOLPIX P100 (10.3 megapixel). The images were acquired with close-ups of the foot, using auto-focus without zoom or macro functions and an aperture setting of f/2.8 at a distance of around 30–40 cm with the parallel orientation to the plane of an ulcer. The use of flash as the primary light source was avoided, with room lights used instead to ensure consistent colours in the resulting photographs. The images were acquired by medical photographers with specialization in the diabetic foot, all with more than 5 years professional experience in podiatry. As a pre-processing stage, we discarded photographs that exhibited poor focus quality. We also excluded duplicates, identified by hash value for each file.
The DFUC 2020 dataset consists of 4,000 images, with 2,000 used for the training set and 2,000 used for the testing set. An additional 200 images were used for sanity checking; images that DFUC 2020 participants could use to perform initial experiments on their models before the release of the testing set. The training set consists of DFU images only, and the testing set comprised of images of DFU, other foot/skin conditions and images of healthy feet. The dataset is heterogeneous, with aspects such as distance, angle, orientation, lighting, focus and the presence of background objects all varying between photographs. We consider this element of the dataset to be important, given that future models will need to account for numerous environmental factors in a system being used in non-medical settings. The images were captured during regular patient appointments at Lancashire Teaching Hospitals foot clinics; therefore, some images were taken from the same subjects at different intervals. Thus, the same ulcer may be present in the dataset more than once, but at different stages of development, at different angles and lighting conditions.
The following describes other notable elements of the dataset, where a case refers to a single image.
- Cases may exhibit more than one DFU.
- Cases exhibit DFU at different stages of healing.
- Cases may not always show all of the foot.
- Cases may show one or two feet, although there may not always be a DFU on each foot.
- Cases may exhibit partial amputations of the foot.
- Cases may exhibit deformity of the foot of varying degrees (Charcot arthropathy).
- Cases may exhibit background objects, such as medical equipment, doctor’s hands, or wound dressings, but no identifiable patient information.
- Cases may exhibit partial blurring.
- Cases may exhibit partial obfuscation of the wound by medical instruments.
- Cases may exhibit signs of debridement, the area of which is often much larger than the ulcer itself.
- Cases may exhibit the presence of all or part of a toenail within a bounding box.
- Cases exhibit subjects of a variety of ethnicities – training set: 1,987 white, 13 non-white; testing set: 1,938 white, 62 non-white; sanity-check set: 194 white; 6 non-white.
- Cases may exhibit signs of infection and/or ischaemia.
- A small number of cases may exhibit the patient’s face. In these instances, the face has been blurred to protect patient identity.
- Cases may exhibit a time stamp printed on the image. If a DFU is obfuscated by a time stamp, the bounding box was adjusted to
include as much of the wound as possible, while excluding the time stamp. - Cases may exhibit imprint patterns resulting from close contact with wound dressings.
- Cases may exhibit unmarked circular stickers or rulers placed close to the wound area, used as a reference point for wound size measurement. Bounding boxes were adjusted to exclude rulers.
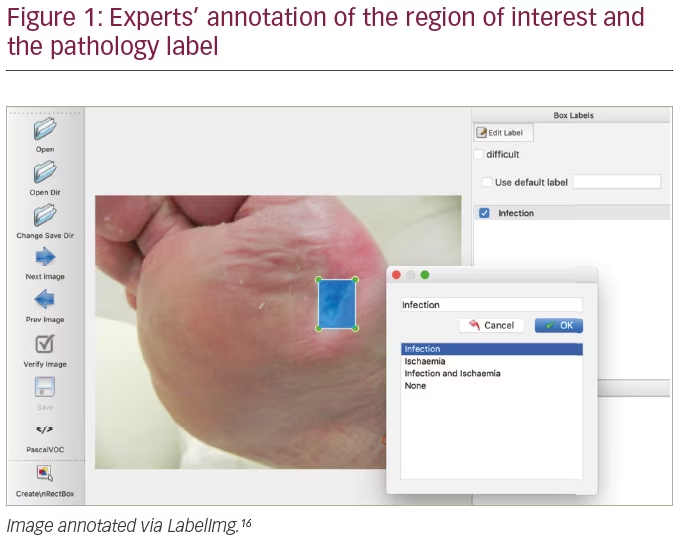
All training, validation and test cases were annotated with the location of foot ulcers in xmin, ymin, xmax and ymax coordinates (Figure 1). Two annotation tools were used to annotate the images: LabelImg16 and VGG Image Annotator.17 These were used to annotate images with a bounding box indicating the ulcer location. The ground truth was produced by three healthcare professionals who specialize in treating diabetic foot ulcers and associated pathology (two podiatrists and a consultant physician with specialization in the diabetic foot, all with more than 5 years professional experience). The instruction for annotation was to label each ulcer with a bounding box. If there was disagreement on DFU annotations, the final decision was mutually settled with the consent of all.
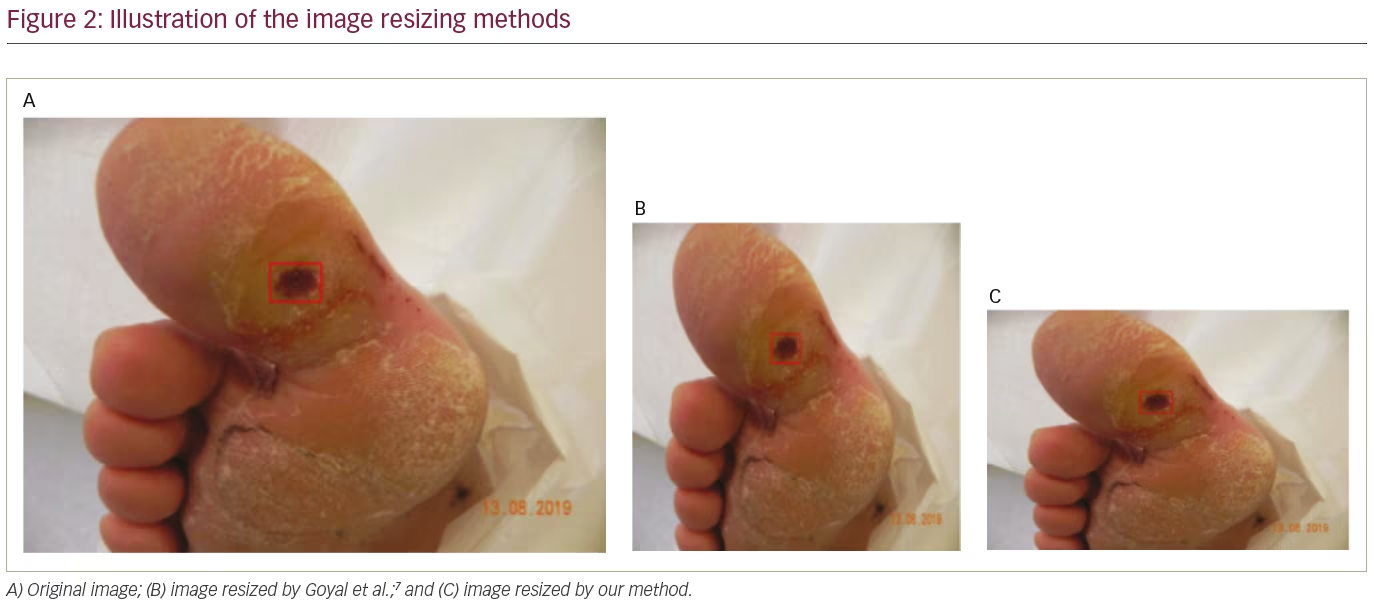
In this dataset, the size of foot images varied between 1,600 x 1,200 and 3,648 x 2,736 pixels. For the release dataset, we resized all images to 640 x 480 pixels to reduce computational costs during training. Unlike the approach by Goyal et al.,7 we preserved the aspect ratio of the images using the high-quality anti-alias down-sampling filter method found in the Python Imaging Library.18 Figure 2A shows the original image with ground truth annotation. Figure 2B shows the resized image by Goyal et al.,7 where the ulcer size and shape changed. We maintained the aspect ratio while resizing, as illustrated in Figure 2C.
Benchmark algorithms
To benchmark predictive performance on the dataset, we conducted experiments with three popular deep learning object detection networks: faster R-CNN,19 You Only Look Once (YOLO) version 5,20 and EfficientDet.12,21 Each of these networks is described as follows.
Faster R-CNN was introduced by Ren et al.19 This network is comprised of three sub-networks: a feature extraction network, a region proposal network (RPN) and a detection network (R-CNN). The feature network extracts features from an image that are then passed to the RPN which uses selective search to generate a series of proposals. Selective search uses a hierarchical grouping algorithm to group similar regions using size, shape and texture.22 These proposals represent locations where objects (of any type) have been initially detected (regions of interest). The outputs from both the feature network and the RPN are then passed to the detection network, which further refines the RPN output and generates the bounding boxes for detected objects. Non-maximum suppression and bounding box regression are used to eliminate duplicate detections and to optimize bounding box positions.23
YOLO was introduced by Redmon et al.24 The authors focused on speed and real-time object detection. Since then, YOLO has become widely used in object detection with the latest versions being YOLOv425 and YOLOv5, produced by other authors. YOLOv5 requires an image to be passed through the network only once. A data loader is used for automatic data augmentation in three stages: (1) scaling, (2) colour space adjustment, and (3) mosaic augmentation. Mosaic augmentation combines four images into four tiles of random ratios, and helps to overcome the limitation of older YOLO networks’ ability to detect smaller objects. A single convolutional neural network is used to process multiple predictions and class probabilities. Non-maximum suppression is used to ensure that each object in an image is only detected once.26
EfficientNet (classification) and EfficientDet (object detection) were introduced by Tan et al.12,27 EfficientDet applies feature fusion to combine representations of an image at different resolutions. Learnable weights are applied at this stage so the network can determine which combinations contribute to the most confident predictions. The final stage uses the feature network outputs to predict class and to plot bounding box positions. EfficientDet is highly scalable, allowing all three sub-networks (and image resolution) to be jointly scaled. This allows the network to be tuned for different target hardware platforms to accommodate variations in hardware capability.12,28
Assessment methods
To enable a fair technical comparison in the DFUC 2020 challenge, participants were not permitted to use external training data unless they agreed that it could be shared with the research community. Participants were also encouraged to report the effect of using a larger training dataset on their techniques.
For performance metrics, F1 score and mAP are used to assess the predictive performance of each detection model that has been trained using the training dataset. Participants were required to record all their detections (including multiple detections) in a log file. A true positive is obtained when the intersection over union (IoU) of the bounding box is greater or equal to 0.5, which is defined by:
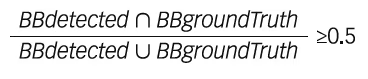
where BBgroundTruth is the bounding box provided by the experts on ulcer location, and BBdetected is the bounding box detected by the algorithm.
F1 score is the harmonic mean of precision and recall, and provides a more suitable measure of predictive performance than the plain percentage of correct predictions in this application. F1 score is used, as false negatives and false positives are crucial, while the number of true negatives can be considered less important. False positives will result in additional cost and time burden to foot clinics, while false negatives will risk further foot complications. The relevant mathematical expressions are as follows:
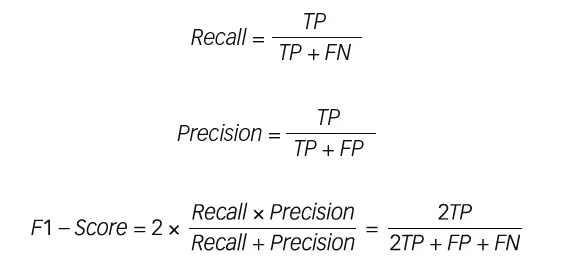
where TP is the total number of true positives, FP is the total number of false positives and FN is the total number of false negatives.
In the field of object detection, mAP is a widely accepted performance metric. This metric is used extensively to measure the overlap percentage of the prediction made by the model and ground truth,2 and is defined as the average of average precision for all classes:
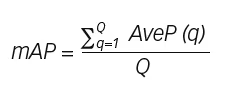
where a class represents the occurrence of a DFU, Q is the number of queries in the set (testing set images), and AveP(q) is the average precision for a given query, q. The exact method of mAP calculation can vary between networks and datasets, often depending on the size of the object that the network has been trained to identify. The definition in the final equation was deemed suitable for the DFUC 2020 challenge since the size of DFUs in the dataset was not constant.
All missing results, e.g. images with no labelled coordinates, are treated as if no DFU had been detected on the image. We evaluated the performance of the baseline algorithms without any post-processing. First, we compared the precision, recall, F1 score and mAP of the baseline algorithms at IoU ≥0.5; we then compared the mAP at IoUs of 0.5–0.9, with an increment of 0.1.
Benchmark experiments
For faster R-CNN, we assessed the performance of three different deep learning network backbone architectures: ResNet101 (residual neural network 101), Inception-v2-ResNet101 and R-FCN (region-based fully convolutional network). For the experimental settings, we used a batch size of 2 and ran gradient descent for 100 epochs initially to observe the loss. We began with a learning rate of 0.002, then reduced this to 0.0002 in epoch 40, and subsequently to 0.00002 in epoch 60. We trained the models for 60 epochs as this was the point at which we observed network convergence.
For the EfficientDet experiment, training was completed using Adam stochastic optimization, with a batch size of 32 for 50 epochs (1,000 steps per epoch) and a learning rate of 0.001. The EfficientNet-B0 network architecture, pretrained on ImageNet, was used as the backbone during training. Random transforms were used as a pre-processing stage to provide automatic data augmentation. To benchmark the performance of the YOLO network on our dataset, we implemented YOLOv5. This is due to the simplicity in installation and superior training and inference times compared to older versions of the network. For our implementation, we used a batch size of 8, and a pre-trained model from MS COCO YOLOv5s provided by the originator of YOLOv5.20
The system configuration used for the R-FCN, faster R-CNN ResNet-101, faster R-CNN Inception-v2-ResNet101 and YOLOv5 experiments were: (1) hardware: CPU – Intel i7-6700 at 4.00Ghz, GPU – NVIDIA TITAN X 12Gb, RAM – 32GB DDR4 (2) software: Ubuntu Linux 16.04 and Tensorflow. The system configuration used for the EfficientDet experiment was: (1) hardware: CPU – Intel i7-8700 at 4.6Ghz, GPU – EVGA GTX 1080 Ti SC 11GB GDDR5X, RAM – 16GB DDR4 (2) software: Ubuntu Linux 20.04 LTS with Keras and Tensorflow.
Results
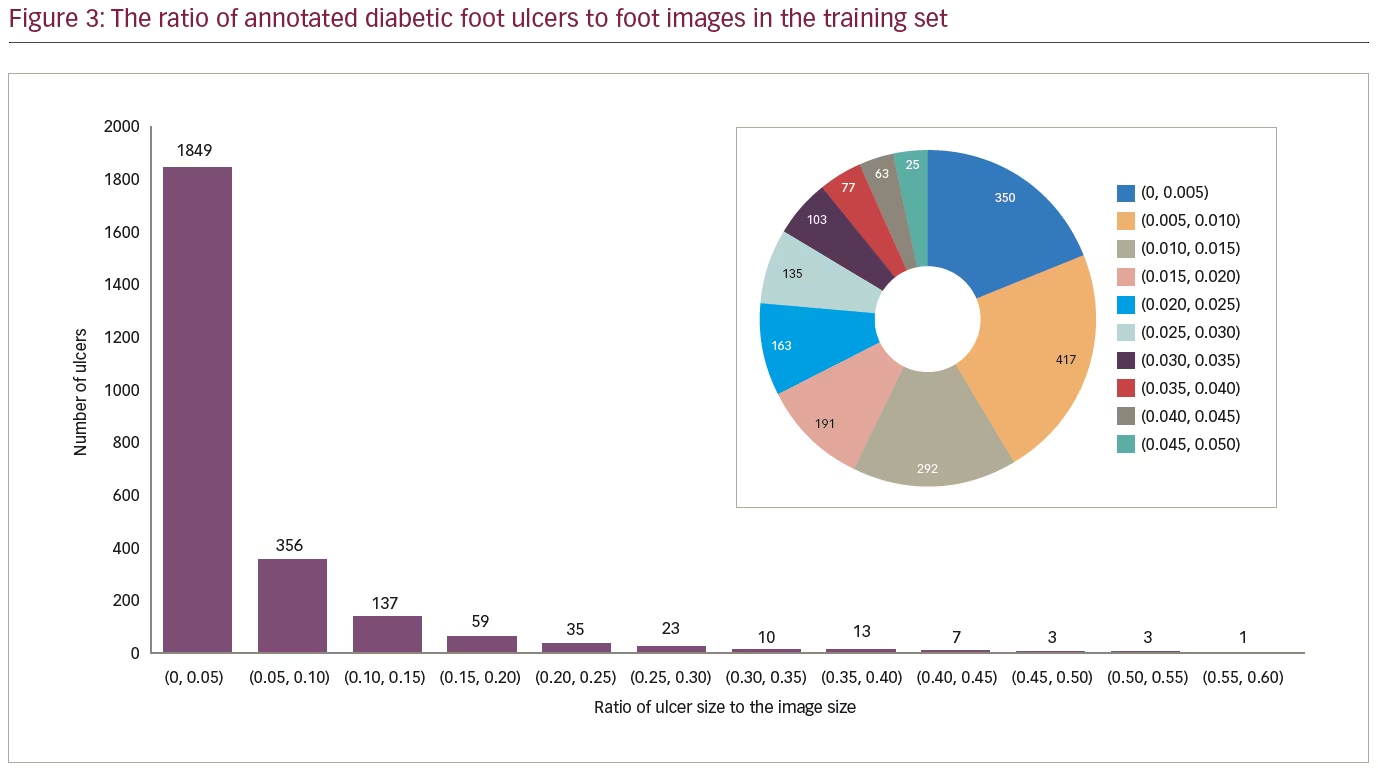
For the training set, there were a total of 2,496 ulcers. A number of images exhibited more than one foot, or more than one ulcer, hence the discrepancy between the number of images and the number of ulcers. The size distribution of the ulcers in proportion to the foot image size is presented in Figure 3. We observed that the size for the majority of ulcers (1,849 images, 74.08%) was <5% of the image size, indicating that the size of ulcers was relatively small. When conducting further analyses on these images, we found that the majority of ulcers (1,250 images, 50.08%) were <2% of the image size.
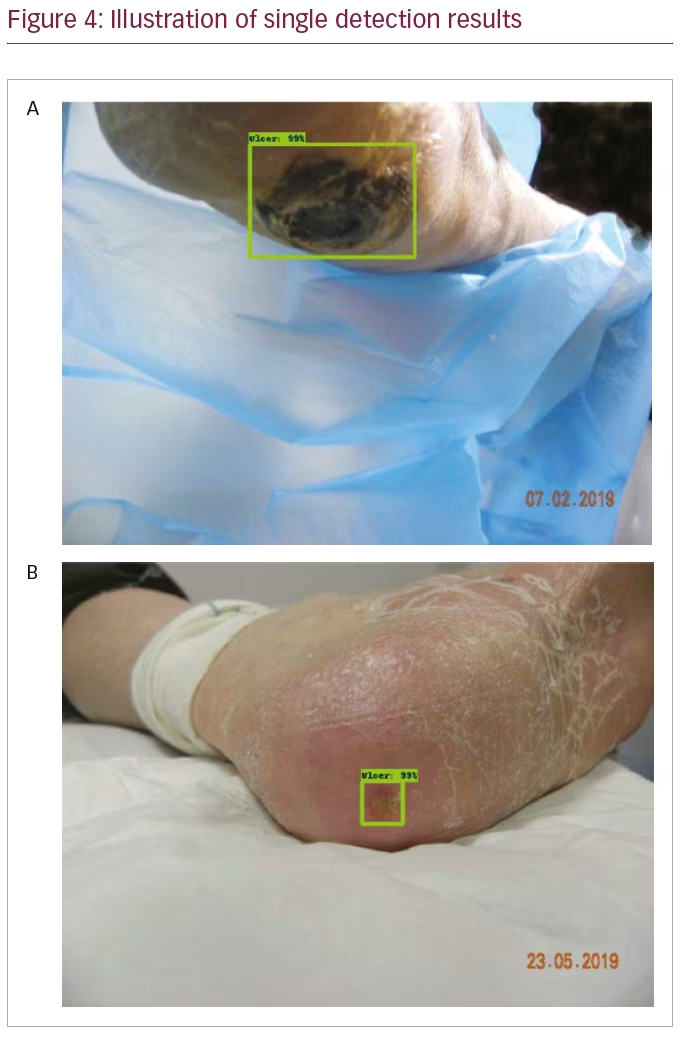
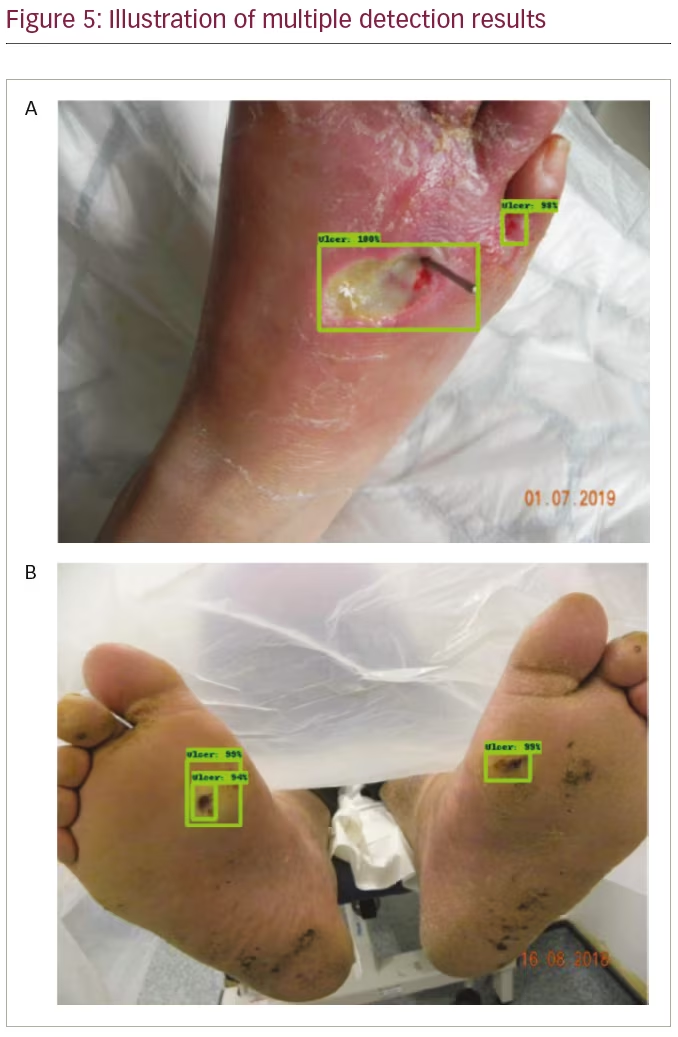
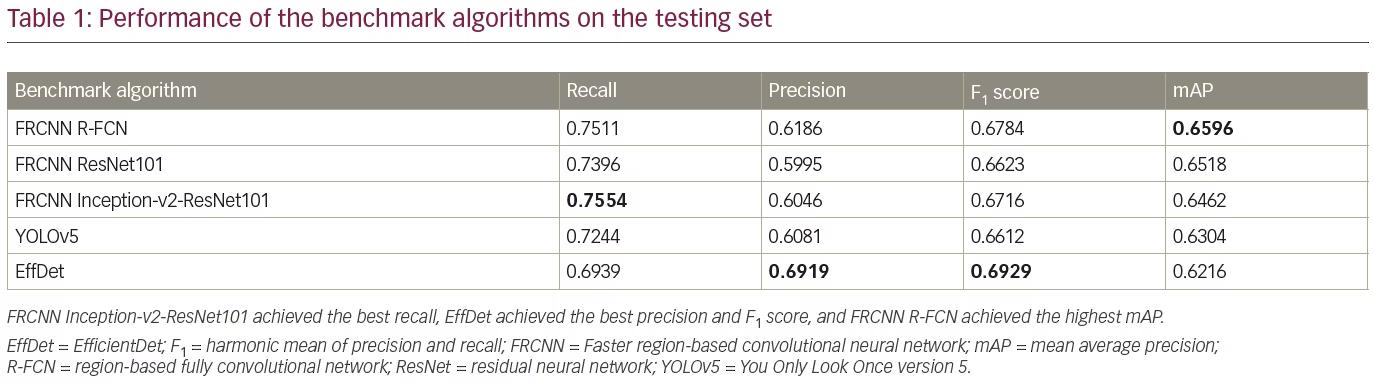
The trained detection models detected single regions with high confidence, as illustrated in Figure 4. Additionally, each trained model detected multiple regions, as illustrated in Figure 5. Table 1 compares the performance of the benchmark algorithms in recall, precision, F1 score and mAP. The faster R-CNN networks achieved high recall, with faster R-CNN Inception-v2-ResNet101 achieving the best result of 0.7554. However, the precision is lower compared with other networks due to the high number of false positives. EfficientDet has the best precision of 0.6919, which is comparable with its recall of 0.6939. When comparing the F1 score, EfficientDet achieved the best result (0.6929). However, it had the lowest mAP (0.6216). The faster R-CNN networks achieved higher mAP, with the best result of 0.6596 achieved by faster R-CNN R-FCN.
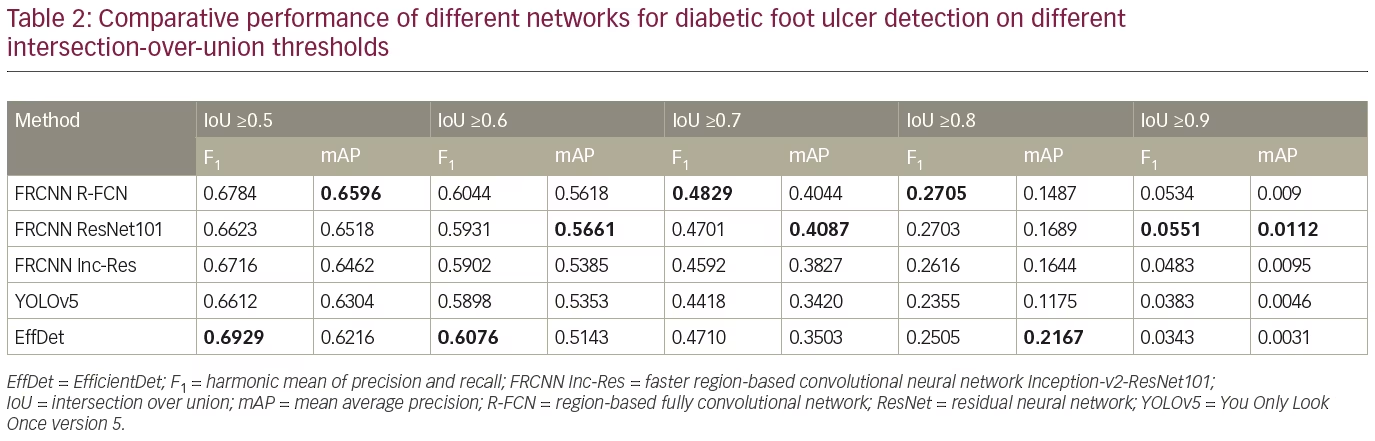
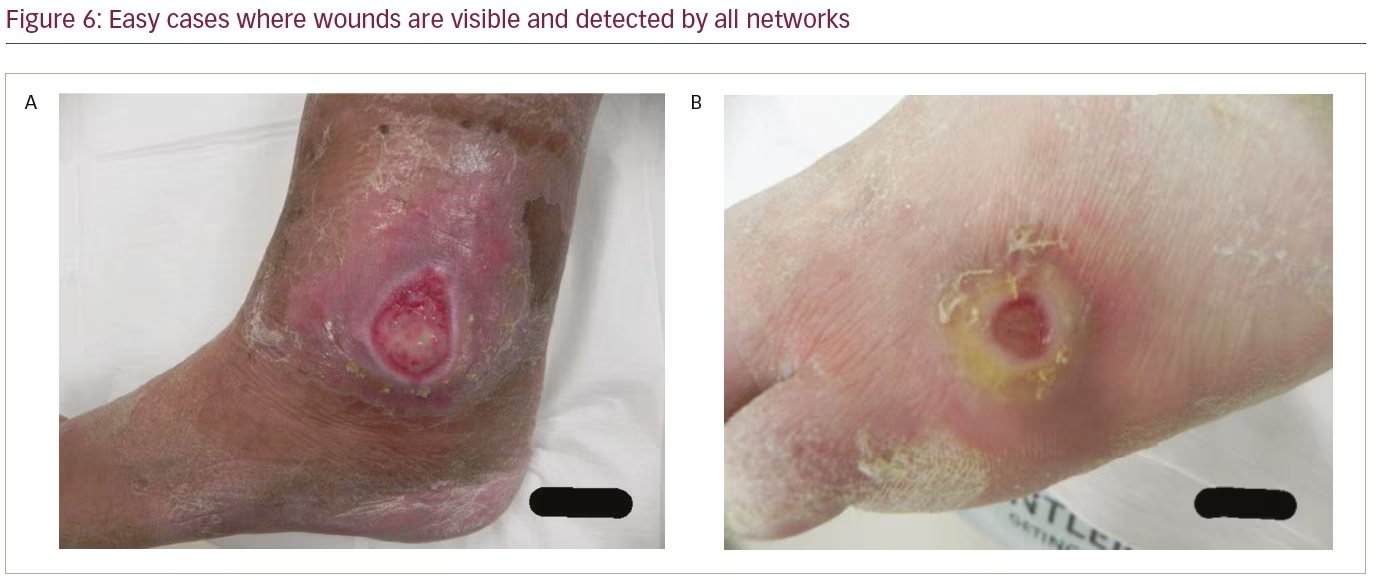
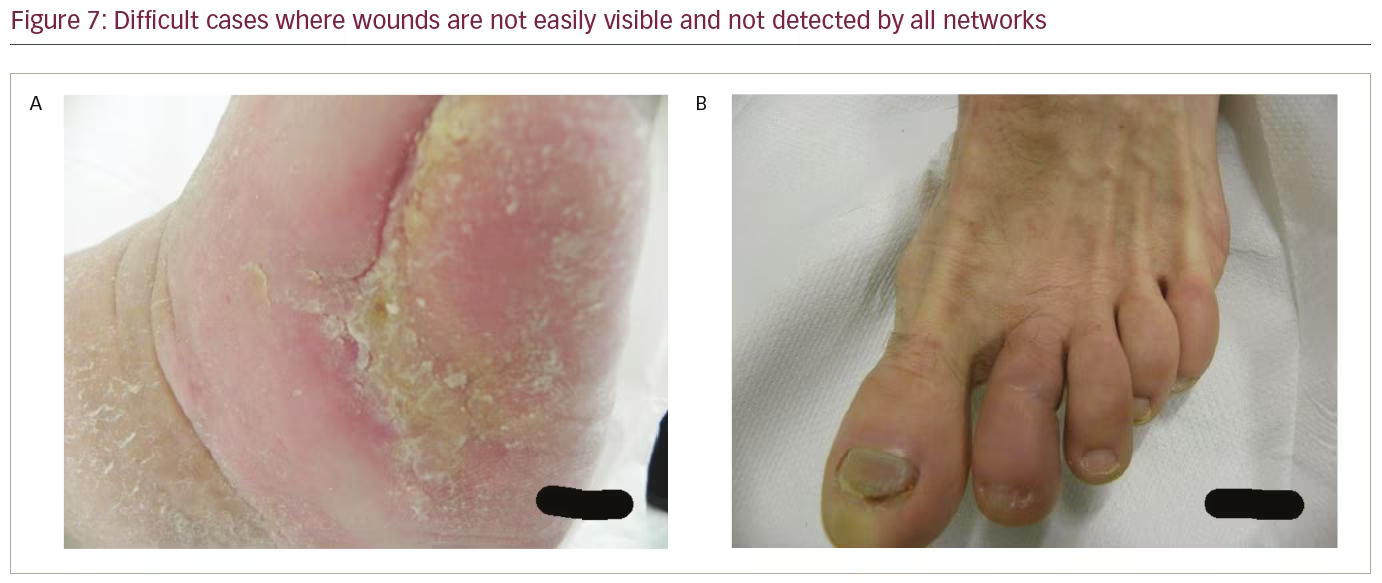
To further analyze the results, Table 2 compares the performance of the networks on different IoU thresholds, from 0.5 to 0.9 with an increment of 0.1. While other networks achieved better recall at 0.5 IoU, EfficientDet shows a better trade-off between recall and precision, which yields the best F1 score. In general, faster R-CNN networks achieved better mAP at 0.5 and remain the leader for mAP at 0.6 to mAP at 0.9 as shown in Table 2. It is also noted that the F1 score for faster R-CNN is better than that of EfficientDet at 0.7 onwards. Figure 6 shows two easy cases detected by all networks, while Figure 7 shows two difficult cases that were missed by all networks.
Discussion
In this paper, we present the largest publicly available DFU dataset together with baseline results generated using three popular deep-learning object-detection networks that were trained using the dataset. No manual pre-processing, fine-tuning or post-processing steps were used beyond those already implemented by each network. We observed that the networks achieved comparable results. Superior results may be achievable by using different anchor settings, for example, with YOLOv5, or by automated removal of duplicate detections.
Non-DFU images were included in our testing dataset to challenge the ability of each network. These images show various skin conditions on different regions of the body, included images of keloids, onychomycosis and psoriasis, many of which share common visual traits with DFU. For the development of future models, we will add images of non-DFU conditions into a second classifier so that the model is more robust. Future work will assess the efficacy of the other available EfficientDet backbones on our dataset. We will also investigate the ability of generative adversarial networks to generate convincing images of DFUs that could be used as data augmentation. We also acknowledge that there is a bias in the dataset, given that the vast majority of subjects are white. We intend to address this issue in future work by working with international collaborators to obtain images that exhibit a variety of skin tones.
Conclusion
This paper presents the largest DFU dataset made publicly available for the research community. The dataset was assembled for the DFUC 2020 challenge, held in conjunction with the MICCAI 2020 conference, and we report baseline results for the DFU test set using state-of-the-art object detection algorithms. The dataset will continue to be available for research after the challenge, in order to motivate algorithm development in this domain. Additionally, we will report the results of the challenge in the near future. For our longer-term plan, we will continue to collect and annotate DFU image data.